Introduction
Ever wondered how to make your code run faster, especially when dealing with large datasets? Enter SIMD (Single Instruction, Multiple Data), a powerful technique that can significantly boost your program's performance by processing multiple data points simultaneously. In this blog post, we'll dive into what SIMD is, the problems it solves, how it works under the hood, and how you can use it in C++ and Python. We'll also explore its advantages, disadvantages, and some notable frameworks and libraries that leverage SIMD. Let's get started!
The Problem SIMD Solves
Traditional scalar processors execute one instruction on one data point at a time. This approach can be inefficient for tasks that involve repetitive operations on large datasets, such as image processing, audio processing, and numerical simulations. SIMD addresses this inefficiency by allowing a single instruction to operate on multiple data points simultaneously, significantly speeding up the computation.
How SIMD Solves the Problem
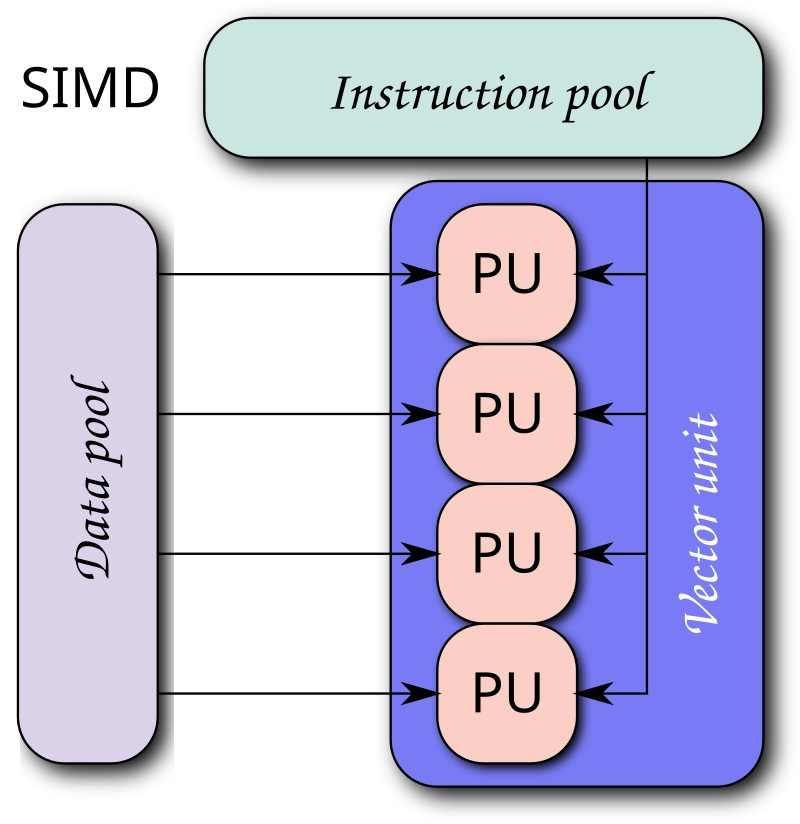
SIMD achieves parallelism by using vector registers and vector instructions. Instead of processing one data element at a time, SIMD instructions operate on vectors, which are arrays of data elements. This parallel processing capability is particularly beneficial for tasks that involve the same operation on multiple data points.
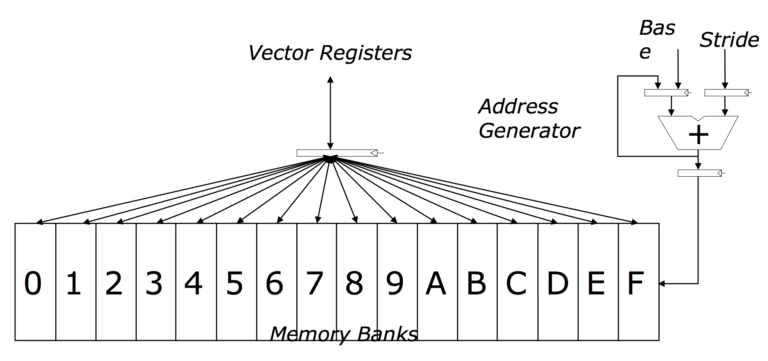
Vector Registers
Vector registers are special hardware registers in the CPU designed to hold multiple data elements. For example, a 256-bit vector register can hold eight 32-bit floating-point numbers. These registers enable the simultaneous execution of operations on multiple data elements, making SIMD highly efficient for data-parallel tasks.
Vector Instructions
Vector instructions are specialized CPU instructions that operate on vector registers. These instructions can perform operations like addition, subtraction, multiplication, and more on all elements of the vector registers in a single instruction cycle. This parallelism is what gives SIMD its performance boost.
Here's a simple illustration to help visualize vector registers and SIMD:
Vector registers | attribution: http://thebeardsage.com/vector-architecture/
SIMD | attribution: By Vadikus - Own work, CC BY-SA 4.0, https://commons.wikimedia.org/w/index.php?curid=39715273
Using SIMD in C++
C++ provides several ways to leverage SIMD instructions, including compiler intrinsics and libraries like Intel's Integrated Performance Primitives (IPP) and the SIMD wrapper library, Eigen.
Example: Using SIMD with Compiler Intrinsics
Here's a simple example of using SIMD intrinsics in C++ to add two arrays of floats:
#include <immintrin.h>
#include <iostream>
// Function to add two arrays using SIMD intrinsics
void add_arrays(const float* a, const float* b, float* result, size_t size) {
size_t i;
// Loop through the arrays in chunks of 8 (since we're using 256-bit registers)
for (i = 0; i < size; i += 8) {
// Load 8 floats from each array into SIMD registers
__m256 vec_a = _mm256_loadu_ps(&a[i]);
__m256 vec_b = _mm256_loadu_ps(&b[i]);
// Perform element-wise addition of the two SIMD registers
__m256 vec_result = _mm256_add_ps(vec_a, vec_b);
// Store the result back into the result array
_mm256_storeu_ps(&result[i], vec_result);
}
}
int main() {
const size_t size = 16;
// Initialize two arrays with sample data
float a[size] = {1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, 13.0, 14.0, 15.0, 16.0};
float b[size] = {16.0, 15.0, 14.0, 13.0, 12.0, 11.0, 10.0, 9.0, 8.0, 7.0, 6.0, 5.0, 4.0, 3.0, 2.0, 1.0};
float result[size];
// Call the SIMD addition function
add_arrays(a, b, result, size);
// Print the result array
for (size_t i = 0; i < size; ++i) {
std::cout << result[i] << " ";
}
std::cout << std::endl;
return 0;
}
Example: Using SIMD with Eigen Library
Eigen is a C++ template library for linear algebra, including matrices, vectors, numerical solvers, and related algorithms. The Dense module provides functionalities for dense matrices and arrays, which are commonly used in linear algebra operations. Here's an example of using Eigen to add two vectors:
#include <Eigen/Dense>
#include <iostream>
int main() {
Eigen::VectorXf a(8);
Eigen::VectorXf b(8);
a << 1, 2, 3, 4, 5, 6, 7, 8;
b << 8, 7, 6, 5, 4, 3, 2, 1;
Eigen::VectorXf result = a + b;
std::cout << "Result: " << result.transpose() << std::endl;
return 0;
}
Using SIMD in Python
Python, being an interpreted language, doesn't natively support SIMD instructions. However, libraries like NumPy and Numba can leverage SIMD under the hood to optimize performance.
Example: Using SIMD with NumPy
NumPy is a powerful library for numerical computing in Python. It uses optimized C and Fortran libraries that can take advantage of SIMD instructions.
import numpy as np
a = np.array([1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0], dtype=np.float32)
b = np.array([8.0, 7.0, 6.0, 5.0, 4.0, 3.0, 2.0, 1.0], dtype=np.float32)
result = a + b
print("Result:", result)
Example: Using SIMD with Numba
Numba is a JIT compiler for Python that can optimize numerical functions to use SIMD instructions.
import numpy as np
from numba import vectorize
@vectorize(['float32(float32, float32)'], target='parallel')
def add_arrays(a, b):
return a + b
a = np.array([1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0], dtype=np.float32)
b = np.array([8.0, 7.0, 6.0, 5.0, 4.0, 3.0, 2.0, 1.0], dtype=np.float32)
result = add_arrays(a, b)
print("Result:", result)
Performance Benchmarks
To see the performance benefits of SIMD, I ran some benchmarks comparing the SIMD and non-SIMD versions of the Python code. Here's what I found:
- SIMD Time: 0.00175 seconds
- Non-SIMD Time: 0.08446 seconds
- Speedup: ~48.15x
Here's the code used for benchmarking:
import numpy as np
from numba import vectorize
import time
# Define the size of the arrays
size = 1000000
# Create two large arrays of random floats
a = np.random.rand(size).astype(np.float32)
b = np.random.rand(size).astype(np.float32)
# Define the SIMD function using Numba
@vectorize(['float32(float32, float32)'], target='parallel')
def add_arrays(a, b):
return a + b
# Define the non-SIMD function
def add_arrays_non_simd(a, b):
return a + b
# Measure the time taken by the SIMD function
start_time = time.time()
result_simd = add_arrays(a, b)
simd_time = time.time() - start_time
# Measure the time taken by the non-SIMD function
start_time = time.time()
result_non_simd = add_arrays_non_simd(a, b)
non_simd_time = time.time() - start_time
print(f"SIMD Time: {simd_time} seconds")
print(f"Non-SIMD Time: {non_simd_time} seconds")
print(f"Speedup: {non_simd_time / simd_time}x")
SIMD's Impact on Machine Learning, LLM, and Generative AI
SIMD can have a significant impact on the performance of machine learning (ML), large language models (LLM), and generative AI applications. These fields often involve processing large datasets and performing repetitive operations, making them ideal candidates for SIMD optimization.
- Machine Learning: SIMD can speed up matrix operations, which are fundamental to training and inference in ML models. Faster computations lead to shorter training times and more efficient inference.
- Large Language Models (LLM): LLMs, like GPT-3, involve extensive matrix multiplications and other linear algebra operations. SIMD can accelerate these operations, improving the model's performance and reducing latency.
- Generative AI: Generative models, such as GANs and VAEs, benefit from SIMD by speeding up the training process and enabling real-time generation of high-quality content.
By leveraging SIMD, developers can achieve significant performance gains in these computationally intensive fields, leading to faster and more efficient AI applications.
Advantages of SIMD
- Performance: SIMD can significantly speed up computations by processing multiple data points simultaneously.
- Efficiency: SIMD reduces the number of instructions executed, leading to more efficient use of CPU resources.
- Scalability: SIMD can be scaled to handle larger datasets by increasing the width of vector registers.
Disadvantages of SIMD
- Complexity: Writing SIMD code can be complex and requires a good understanding of the underlying hardware.
- Portability: SIMD code may not be portable across different CPU architectures due to varying SIMD instruction sets.
- Limited Applicability: SIMD is most effective for data-parallel tasks and may not provide benefits for other types of computations.
Considerations
- Alignment: Data alignment is crucial for optimal SIMD performance. Misaligned data can lead to performance penalties.
- Data Size: SIMD is most effective for large datasets where the overhead of setting up SIMD operations is amortized over many data points.
- Instruction Set: Different CPUs support different SIMD instruction sets (e.g., SSE, AVX, NEON). Ensure your code targets the appropriate instruction set for your hardware.
Notable Frameworks and Libraries
- Intel Integrated Performance Primitives (IPP): A library of highly optimized functions for multimedia, data processing, and communications applications.
- Eigen: A C++ template library for linear algebra that supports SIMD operations.
- NumPy: A powerful library for numerical computing in Python that leverages SIMD under the hood.
- Numba: A JIT compiler for Python that can optimize numerical functions to use SIMD instructions.
Conclusion
SIMD instructions provide a powerful way to enhance the performance of data-parallel tasks by processing multiple data points simultaneously. While SIMD can be complex to implement, the performance benefits it offers make it a valuable tool for optimizing computationally intensive applications. By leveraging libraries and frameworks that support SIMD, developers can take advantage of this technology without delving into the intricacies of SIMD programming.
References
- SIMDe GitHub Repository: A portable implementation of SIMD intrinsics
- xsimd GitHub Repository: A C++ wrapper for SIMD intrinsics
- VCL GitHub Repository: A library for using SIMD vector classes in C++
- simdjson GitHub Repository: Parsing gigabytes of JSON per second
- GCC SIMD Documentation: Documentation on using SIMD with GCC



Top comments (0)