Introduction
Uniform Manifold Approximation and Projection (UMAP) is a well-known dimensionality reduction method along with t-SNE.
Ruby users often use Rumale for machine learning. t-SNE is included in Rumale, but UMAP is not.
I have created a Ruby binding for Umappp, a C++ library, and will post it here before I forget.
GitHub: https://github.com/kojix2/ruby-umappp
Create bindings when Ruby libraries do not exist!
Since the Ruby language is a relatively minor language in the field of data analysis, it is often the case that a library that implements what you want to do does not exist. In such cases, you can look for libraries in languages such as C or Rust to build Ruby bindings. GitHub allows you to search for code by language. This can be used to find libraries for C. Since GitHub allows tagging of projects, searching for the target tag can also be helpful. However, UMAP seems to be difficult to implement, and I could not find a C library that implements UMAP. Instead, I found a library that implements UMAP in C++. That is Umappp.
Umappp - UMAP C++ implementation
Umappp is a C++ library implemented by Aaron Lun. it is developed based on the R library uwot. it is implemented in C++ and uses OpenMP, so high performance is expected.
Calling C++ functions from the Ruby language
Extension libraries using C++ are common in the R language, but C++ extension libraries are not so widely used in the Ruby language. Recently, Rust is very popular, and I sometimes see people building Ruby extensions in Rust, but I have not seen many new C++ extension libraries.
However, creating C++ bindings in Ruby is easier than you might think. I have no experience with C++, but I was able to call C++ from Ruby.
There are two ways to write Ruby extensions in C++. One is Rice and the other is extpp. In this case, I used Rice because I wanted to use numo.hpp to link Numo::NArray and C++.
In order to be able to compile C++ at runtime, Umappp and all C++ files on which Umappp depends are put in the Vendor directory and distributed as a gem.extconf.rb is a script that creates a Makefile. I didn't know how to write this, so I wrote it based on other projects.
Rice - Ruby Interface for C++ Extensions
Rice is a library developed by Paul Brannan, Charlie Savage, Jason Roelofs and others for writing C++ extensions in Ruby. Using Rice, you can define Ruby modules and methods from C++ code as shown below.
#include <rice/rice.hpp>
#include <rice/stl.hpp>
using namespace Rice;
Hash umappp_default_parameters(Object self)
{
Hash d;
// ...
return d;
}
// ...
extern "C" void Init_umappp()
{
Module rb_mUmappp =
define_module("Umappp")
.define_singleton_method("umappp_run", &umappp_run)
.define_singleton_method("umappp_default_parameters", &umappp_default_parameters);
}
Run UMAP in Ruby
Run UMAP on the famous Iris dataset and the MNIST database.
- For visualization, I used GR.rb.
- For dataset fetching, I used red-datasets and its derived library red-datasets-numo-narray.
Iris dataset
Ruby code:
require "umappp"
require "datasets-numo-narray"
require "gr/plot"
iris = Datasets::LIBSVM.new("iris").to_narray
d = iris[true, 1..-1]
l = iris[true, 0]
r = Umappp.run(d)
x = r[true, 0]
y = r[true, 1]
s = [2000] * l.size
GR.scatter(
x, y, s, l,
title: "iris",
colormap: 16,
colorbar: true
)
gets
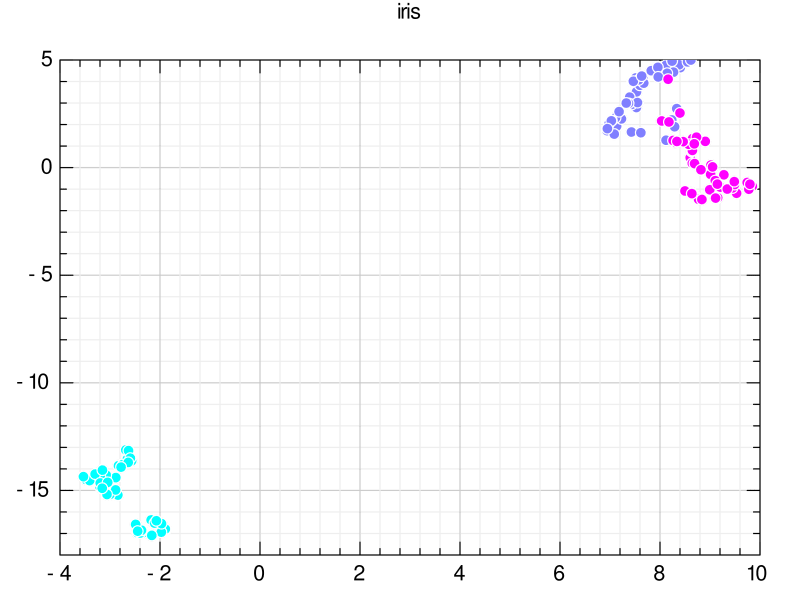
The clusters were clearly separated. Here, label 0 (setosa) is light blue, label 1 (versicolor) is wisteria, and label 2 (virginica) is magenta. The setosa group is clearly separated from the other groups, while versicolor and virginica partially overlap.
Mnist database
Ruby code:
require "umappp"
require "datasets"
require "gr/plot"
require "etc"
mnist = Datasets::MNIST.new
pixels = []
labels = []
mnist.each_with_index do |r, _i|
pixels << r.pixels
labels << r.label
end
puts "start umap"
nproc = Etc.nprocessors
n = nproc > 4 ? nproc - 1 : nproc
d = Umappp.run(pixels, num_threads: n, a: 1.8956, b: 0.8006)
puts "end umap"
x = d[true, 0]
y = d[true, 1]
s = [500] * x.size
GR.scatter(x, y, s, labels, colormap: 0)
gets
If you monitor CPU utilization in an environment with OpenMP installed, you will see that multiple cores are in use.
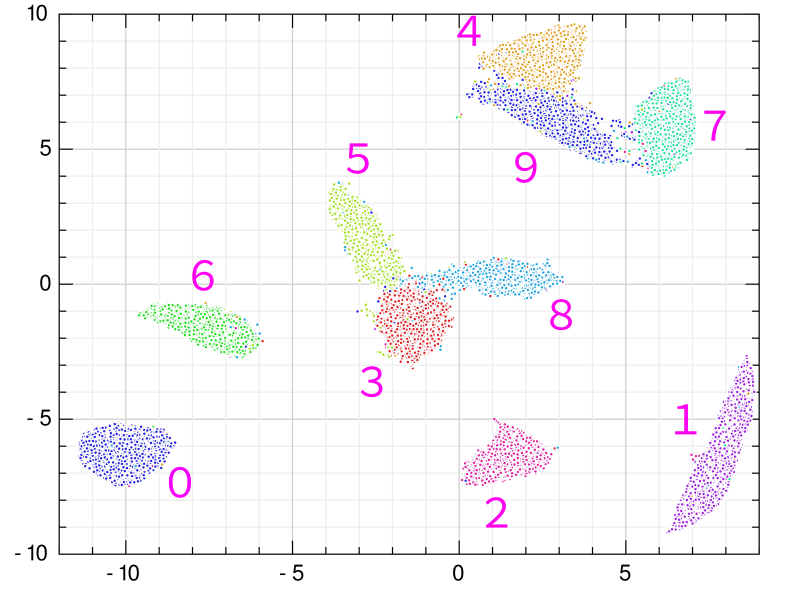
The cluster has been divided very nicely. Each cluster is labeled with a number. This roughly matches the official UMAP results. UMAP is running successfully. 5-3-8 and 4-9-7 form a group. The similarity between the handwritten letters 4 and 9 is intuitively obvious. I have a habit of writing 1 to resemble 7, but not many people do that.
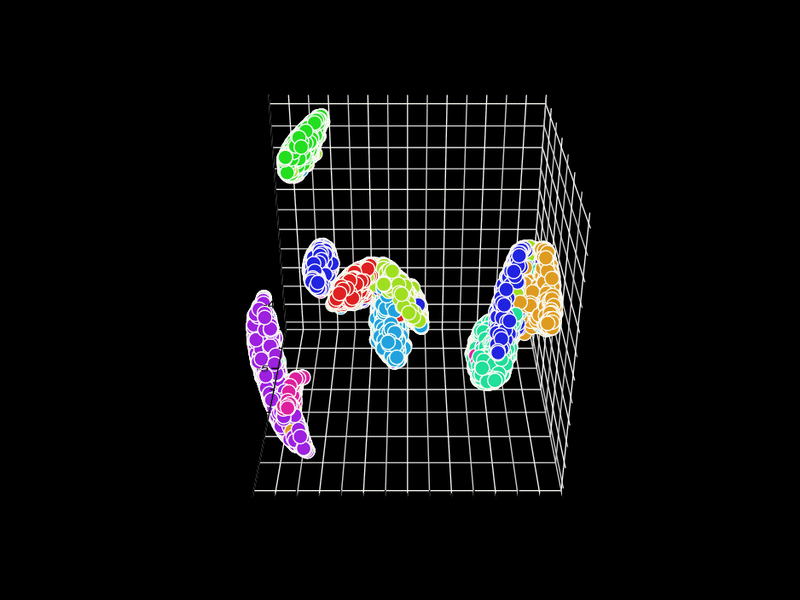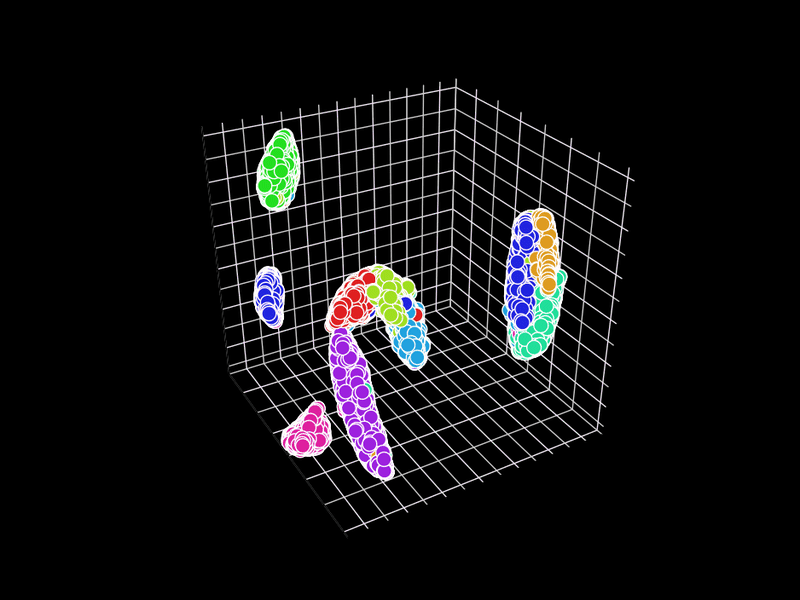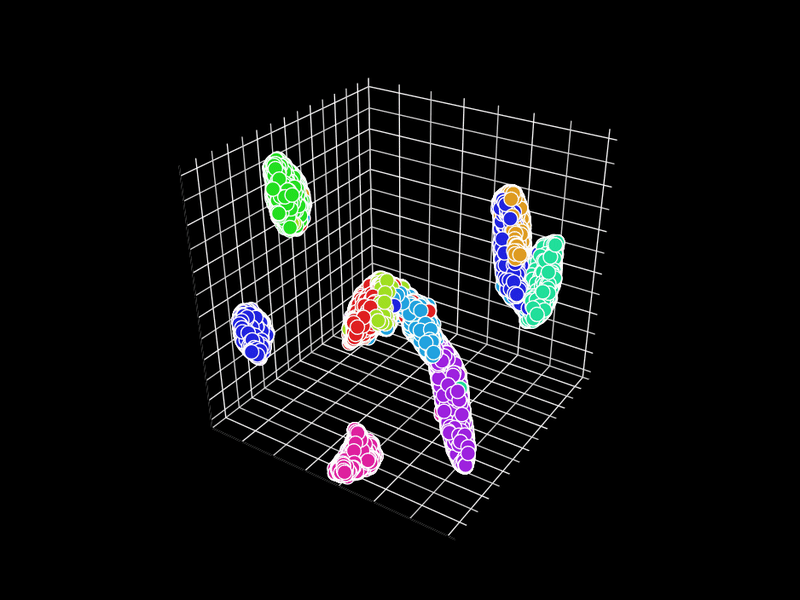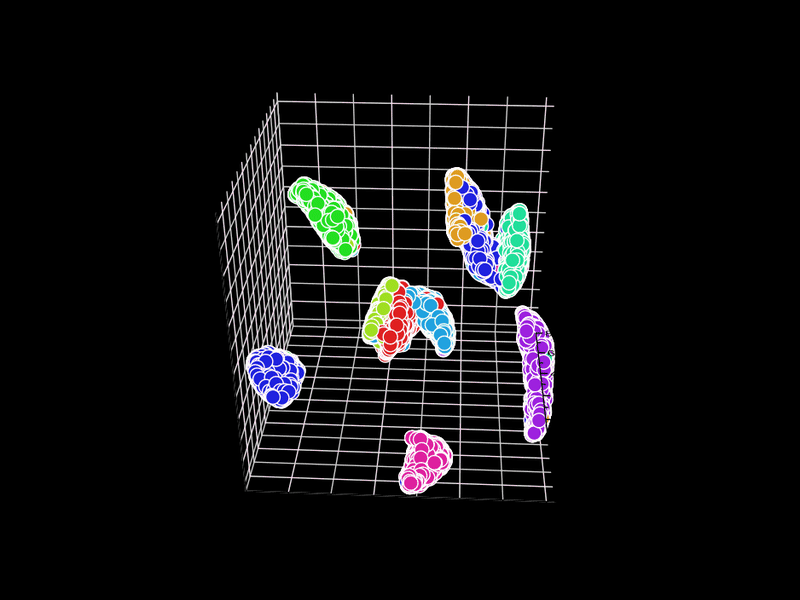
Let's try 3D. GR.rb has a GIF animation output function.
puts "start umap #{n} threads"
d = Umappp.run(pixels, ndim: 3, num_threads: n, a: 1.8956, b: 0.8006)
puts "end umap"
x = d[true, 0]
y = d[true, 1]
z = d[true, 2]
Dir.chdir(__dir__) do
# Save results
File.binwrite("data/mnist2.dat", Marshal.dump([d, labels]))
puts "Saved to data/mnist2.dat"
# Save gif animation
GR.beginprint("data/mnist2.gif") do
30.times do |i|
puts "frame #{i + 1}"
GR.scatter3(x, y, z, labels, colormap: 0, backgroundcolor: 1, rotation: i * 3)
end
end
puts "Saved to data/mnist2.gif"
end
Unfortunately, GR.rb calls polymarker3d in sequence, so some points that should be behind are in front and some points that should be in front are behind. However, it is interesting that the 2D position is consistent with the 3D position.
In GR.rb, it is not clear which set of colors corresponds to which number, since the scatter plot and color bar cannot be displayed at the same time. There is room for improvement on this point.
The author is not very good at mathematics and does not understand the details of how UMAP works at all. So I can't say anything nice about UMAP, but if it works this fast, I thought it might be useful to use UMAP to look up data I'm interested in. I also felt that the results were more reproducible than I expected.
Conclusion
In this post, I have shown how to call C++ from Ruby to execute UMAP. As far as I know, this is the first library that can run UMAP in Ruby, including bindings.
Thus, another Ruby gem has been created.
Have a nice day!




Top comments (0)