Step 1: Data Loading and Initial Analysis
This Python 3 environment comes with many helpful analytics libraries installed
It is defined by the kaggle/python Docker image: https://github.com/kaggle/docker-python
For example, here's several helpful packages to load
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings('ignore')
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk('/kaggle/input'):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the Defects Dataset
train = pd.read_csv('/kaggle/input/train.csv')
test = pd.read_csv('/kaggle/input/test.csv')
# Display first few rows
print("First 5 rows of the training dataset:")
print(train.head())
print("First 5 rows of the test dataset:")
print(test.head())
# Dataset shape
print(f"\nDataset Shape: {train.shape}")
print(f"Number of samples: {train.shape[0]}")
print(f"Number of features: {train.shape[1]}")
print(f"\nDataset Shape: {test.shape}")
print(f"Number of samples: {test.shape[0]}")
print(f"Number of features: {test.shape[1]}")
# Column names and data types
print("\nColumn Information for training dataset:")
print(train.info())
print("\nColumn Information for test datase
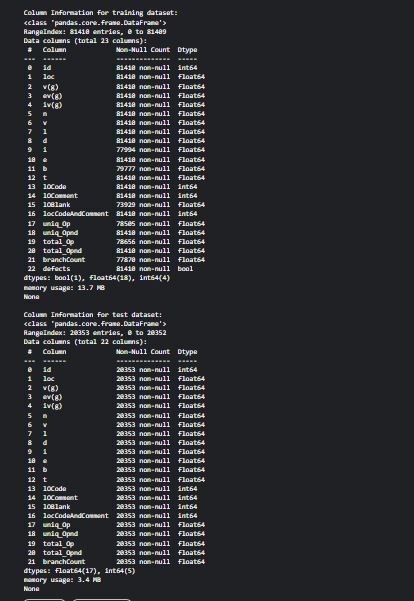t:")
print(test.info())
# Statistical summary
print("Statistical Summary:")
print(train.describe())
#Get the statistical information about the training set
import plotly.express as px
train.describe().T\
.style.bar(subset=['mean'], color=px.colors.qualitative.G10[2])\
.background_gradient(subset=['std'], cmap='Blues')\
.background_gradient(subset=['50%'], cmap='Reds')
# Check for missing values
print("=" * 50)
print("Missing Values Analysis")
print("=" * 50)
missing = train.isnull().sum()
missing_pct = (train.isnull().sum() / len(train)) * 100
missing_train = pd.DataFrame({
'Missing_Count': missing,
'Missing_Percentage': missing_pct
})
print(missing_train[missing_train['Missing_Count'] > 0])
# Check for duplicates
print("=" * 50)
print("Duplicate Analysis")
print("=" * 50)
duplicates = train.duplicated().sum()
print(f"Number of duplicate rows: {duplicates}")
print(f"\nDuplicate rows:")
train[train.duplicated(keep=False)].sort_values('id')
# Check target variable distribution
print("\nTarget Variable Distribution:")
print(train['defects'].value_counts())
print(f"\nClass Balance:")
print(train['defects'].value_counts(normalize=True))
# target value count
defects_count = dict(train['defects'].value_counts())
defects_count
#Check for missing (NaN) value
print(train[train.isna().any(axis=1)])
Step 2: EDA
# visualize data distrubution
fig , axis = plt.subplots(figsize = (25,35))
fig.suptitle('Data Distribution', ha = 'center', fontsize = 20, fontweight = 'bold',y=0.90 )
for idx,col in enumerate(list(train.columns)[1:-1]):
plt.subplot(7,3,idx+1)
ax = sns.histplot(train[col], color = 'blue' ,stat='density' , kde = False,bins = 50)
sns.kdeplot(train[col] , color = 'orange' , ax = ax)
# adjust label names
labels = ['not_defects', 'defects']
# visualize data target
plt.title('Defects Values')
sns.barplot(x = labels ,y = list(defects_count.values()), width = 0.3 , palette = ['blue' , 'orange'])
# visualize correlation
corr = train.corr()
mask = np.triu(np.ones_like(corr))
plt.figure(figsize = (20,10))
plt.title('Correlation Matrix')
sns.heatmap(data = corr , mask = mask , annot = True , cmap = 'coolwarm',annot_kws={"color": "black", "size":9} )
**Step 3 : Data cleaning and preparation**
# convert target to binary encoder
# Encode categorical variables
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
train['defects'] = le.fit_transform(train['defects'])
train.defects.unique()
Handling Missing Values
# Strategy 1: Fill missing values with mean/median
train_cleaned = train.copy()
print("Before filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum())
# Fill numeric missing values with median
#train_cleaned['score'].fillna(train_cleaned['score'].median(), inplace=True)
train_cleaned['i'].fillna(train_cleaned['i'].mean(), inplace=True)
train_cleaned['b'].fillna(train_cleaned['b'].mean(), inplace=True)
train_cleaned['lOBlank'].fillna(train_cleaned['lOBlank'].mean(), inplace=True)
train_cleaned['uniq_Op'].fillna(train_cleaned['uniq_Op'].mean(), inplace=True)
train_cleaned['total_Op'].fillna(train_cleaned['total_Op'].mean(), inplace=True)
train_cleaned['branchCount'].fillna(train_cleaned['branchCount'].mean(), inplace=True)
print("\nAfter filling missing values:")
print(train_cleaned[['i', 'b','lOBlank','uniq_Op','total_Op','branchCount']].isnull().sum())
**Handling Outliers**
# Visualize outliers using box plots
plt.figure(figsize=(12,12))
i=1
for col in train.columns:
plt.subplot(6,6,i)
train[[col]].boxplot()
i+=1
# Detect outliers using IQR method
def detect_outliers_iqr(data, column):
Q1 = data[column].quantile(0.25)
Q3 = data[column].quantile(0.75)
IQR = Q3 - Q1
lower_bound = Q1 - 1.5 * IQR
upper_bound = Q3 + 1.5 * IQR
outliers = data[(data[column] < lower_bound) | (data[column] > upper_bound)]
return outliers, lower_bound, upper_bound
# Detect outliers in age
outliers_b, lower_b, upper_b = detect_outliers_iqr(train_cleaned, 'b')
print(f"b outliers (outside {lower_b:.2f} - {upper_b:.2f}):")
print(outliers_b[['id', 'b']])
#List of numerical columns
num_cols=[col for col in train.columns if (train[col].dtype in ["int64","float64"]) & (train[col].nunique()>50)]
num_cols
#Method to handle outliers
def handle_outliers(
train,
columns,
factor=1.5,
method="clip" # "clip" or "remove"
):
"""
Handle outliers using the IQR method.
Parameters:
train (pd.DataFrame): Input DataFrame
columns (list): List of numeric columns
factor (float): IQR multiplier (default 1.5)
method (str): "clip" to cap values, "remove" to drop rows
Returns:
pd.DataFrame
"""
train = train.copy()
for col in columns:
Q1 = train[col].quantile(0.25)
Q3 = train[col].quantile(0.75)
IQR = Q3 - Q1
lower = Q1 - factor * IQR
upper = Q3 + factor * IQR
if method == "clip":
train[col] = train[col].clip(lower, upper)
elif method == "remove":
train = train[(train[col] >= lower) & (train[col] <= upper)]
else:
raise ValueError("method must be 'clip' or 'remove'")
return train
# Cap outliers
print(f"Before outlier cleaning: {train_cleaned.shape}")
train_cleaned = handle_outliers(train_cleaned, num_cols, method="clip")
print(f"After outlier cleaning: {train_cleaned.shape}")
Step 4 : Model Building and Prediction
# Separate features and target
X= train_cleaned.drop("defects", axis=1)
y= train_cleaned["defects"]
X_train, X_test, y_train, y_test = train_test_split(X, y,test_size=0.1, random_state = 42, stratify=y)
# Initialize Random Forest Classifier
rf_model = RandomForestClassifier(
n_estimators=50,
criterion='gini',
max_depth=20,
min_samples_split=10,
min_samples_leaf=5,
max_features='sqrt',
bootstrap=True,
random_state=42,
n_jobs=-1
)
# Train the model
rf_model.fit(X_train, y_train)
print("Random Forest model trained successfully!")
# Display model parameters
print("\nModel Parameters:")
print(rf_model.get_params())
Model Evaluation
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred)
print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50)
# Initialize Decision Tree Classifier
dt_model = DecisionTreeClassifier(
criterion='gini',
max_depth=5,
min_samples_split=20,
min_samples_leaf=10,
random_state=42
)
# Train the model
dt_model.fit(X_train, y_train)
print("Decision Tree model trained successfully!")
from sklearn.tree import export_text
tree_rules = export_text(dt_model)
print(tree_rules)
# Make predictions
y_train_pred = dt_model.predict(X_train)
y_test_pred = dt_model.predict(X_test)
print("Predictions completed!")
# Calculate accuracy scores
train_accuracy = accuracy_score(y_train, y_train_pred)
test_accuracy = accuracy_score(y_test, y_test_pred)
print("=" * 50)
print("Accuracy Scores")
print("=" * 50)
print(f"Training Accuracy: {train_accuracy:.4f} ({train_accuracy*100:.2f}%)")
print(f"Testing Accuracy: {test_accuracy:.4f} ({test_accuracy*100:.2f}%)")
print("=" * 50)
#linear Regression
def lr(X_train, X_test, y_train, y_test):
model = LinearRegression()
model.fit(X_train, y_train.values.ravel())
y_pred = model.predict(X_test)
error = mean_squared_error(y_test, y_pred)
print(f"Score: {error}")
return model
Submission
#Submission
X_test = test[X_train.columns]
y_test_pred = rf_model.predict(X_test)
submission = pd.DataFrame({
"id": test["id"],
"defects": y_test_pred
})
submission.to_csv("submission.csv", index=False)
submission




















Top comments (0)