Diabetes can be discovered from some parameters well known to the medical community. In this way, in order to help the medical community and computerized systems, especially AI, the National Institute of Diabetes and Digestive and Kidney Diseases published a very useful dataset for training ML algorithms in the detection/prediction of diabetes. This publication can be found on the largest and best known data repository for ML, Kaggle at https://www.kaggle.com/datasets/mathchi/diabetes-data-set.
The diabetes dataset has the following metadata information (source: https://www.kaggle.com/datasets/mathchi/diabetes-data-set):
- Pregnancies: Number of times pregnant
- Glucose: Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- BloodPressure: Diastolic blood pressure (mm Hg)
- SkinThickness: Triceps skin fold thickness (mm)
- Insulin: 2-Hour serum insulin (mu U/ml)
- BMI: Body mass index (weight in kg/(height in m)^2)
- DiabetesPedigreeFunction: Diabetes pedigree function (It provided some data on diabetes mellitus history in relatives and the genetic relationship of those relatives to the patient. This measure of genetic influence gave us an idea of the hereditary risk one might have with the onset of diabetes mellitus - source: https://machinelearningmastery.com/case-study-predicting-the-onset-of-diabetes-within-five-years-part-1-of-3/)
- Age: Age (years)
- Outcome: Class variable (0 or 1)
Number of Instances: 768
Number of Attributes: 8 plus class
For Each Attribute: (all numeric-valued)
- Number of times pregnant
- Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- Diastolic blood pressure (mm Hg)
- Triceps skin fold thickness (mm)
- 2-Hour serum insulin (mu U/ml)
- Body mass index (weight in kg/(height in m)^2)
- Diabetes pedigree function
- Age (years)
- Class variable (0 or 1)
Missing Attribute Values: Yes
Class Distribution: (class value 1 is interpreted as "tested positive for diabetes")
Get the Diabetes data from Kaggle
The Diabetes data from Kaggle can be loaded into an IRIS table using the Health-Dataset application: https://openexchange.intersystems.com/package/Health-Dataset. To do this, from your module.xml project, set the dependency (ModuleReference for Health Dataset):
Module.xml with Health Dataset application reference
<?xml version="1.0" encoding="UTF-8"?>
<Export generator="Cache" version="25">
<Document name="predict-diseases.ZPM">
<Module>
<Name>predict-diseases</Name>
<Version>1.0.0</Version>
<Packaging>module</Packaging>
<SourcesRoot>src/iris</SourcesRoot>
<Resource Name="dc.predict.disease.PKG"/>
<Dependencies>
<ModuleReference>
<Name>swagger-ui</Name>
<Version>1.*.*</Version>
</ModuleReference>
<ModuleReference>
<Name>dataset-health</Name>
<Version>*</Version>
</ModuleReference>
</Dependencies>
<CSPApplication
Url="/predict-diseases"
DispatchClass="dc.predict.disease.PredictDiseaseRESTApp"
MatchRoles=":{$dbrole}"
PasswordAuthEnabled="1"
UnauthenticatedEnabled="1"
Recurse="1"
UseCookies="2"
CookiePath="/predict-diseases"
/>
<CSPApplication
CookiePath="/disease-predictor/"
DefaultTimeout="900"
SourcePath="/src/csp"
DeployPath="${cspdir}/csp/${namespace}/"
MatchRoles=":{$dbrole}"
PasswordAuthEnabled="0"
Recurse="1"
ServeFiles="1"
ServeFilesTimeout="3600"
UnauthenticatedEnabled="1"
Url="/disease-predictor"
UseSessionCookie="2"
/>
</Module>
</Document>
</Export>
Web Frontend and Backend Application to Predict Diabetes
Go to Open Exchange app link (https://openexchange.intersystems.com/package/Disease-Predictor) and follow these steps:
- Clone/git pull the repo into any local directory
$ git clone https://github.com/yurimarx/predict-diseases.git
- Open a Docker terminal in this directory and run:
$ docker-compose build
- Run the IRIS container:
$ docker-compose up -d
- Go to Execute Query into Management Portal to train the AI model: http://localhost:52773/csp/sys/exp/%25CSP.UI.Portal.SQL.Home.zen?$NAMESPACE=USER
- Create the VIEW used to train:
CREATE VIEW DiabetesTrain AS SELECT Outcome, age, bloodpressure, bmi, diabetespedigree, glucose, insulin, pregnancies, skinthickness FROM dc_data_health.Diabetes
- Create the AI Model using the view:
CREATE MODEL DiabetesModel PREDICTING (Outcome) FROM DiabetesTrain
- Train the model:
TRAIN MODEL DiabetesModel
-
Go to http://localhost:52773/disease-predictor/index.html to use the Disease Predictor frontend and predict diseases like this:
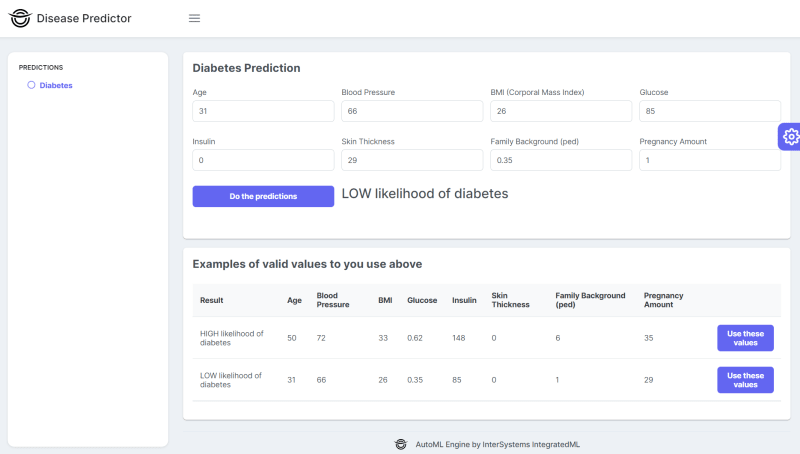
Behind the scenes
Backend ClassMethod to predict Diabetes
InterSystems IRIS allows you execute SELECT to predict using the previous model created.
Backend ClassMethod to predict Diabetes
/// Predict Diabetes
ClassMethod PredictDiabetes() As %Status
{
Try {
Set data = {}.%FromJSON(%request.Content)
Set qry = "SELECT PREDICT(DiabetesModel) As PredictedDiabetes, "
_"age, bloodpressure, bmi, diabetespedigree, glucose, insulin, "
_"pregnancies, skinthickness "
_"FROM (SELECT "_data.age_" AS age, "
_data.bloodpressure_" As bloodpressure, "
_data.bmi_" AS bmi, "
_data.diabetespedigree_" AS diabetespedigree, "
_data.glucose_" As glucose, "
_data.insulin_" AS insulin, "
_data.pregnancies_" As pregnancies, "
_data.skinthickness_" AS skinthickness)"
Set tStatement = ##class(%SQL.Statement).%New()
Set qStatus = tStatement.%Prepare(qry)
If qStatus'=1 {WRITE "%Prepare failed:" DO $System.Status.DisplayError(qStatus) QUIT}
Set rset = tStatement.%Execute()
Do rset.%Next()
Set Response = {}
Set Response.PredictedDiabetes = rset.PredictedDiabetes
Set Response.age = rset.age
Set Response.bloodpressure = rset.bloodpressure
Set Response.bmi = rset.bmi
Set Response.diabetespedigree = rset.diabetespedigree
Set Response.glucose = rset.glucose
Set Response.insulin = rset.insulin
Set Response.pregnancies = rset.pregnancies
Set Response.skinthickness = rset.skinthickness
Set %response.Status = 200
Set %response.Headers("Access-Control-Allow-Origin")="*"
Write Response.%ToJSON()
Return 1
} Catch err {
write !, "Error name: ", ?20, err.Name,
!, "Error code: ", ?20, err.Code,
!, "Error location: ", ?20, err.Location,
!, "Additional data: ", ?20, err.Data, !
Return 0
}
}
Now, any web application can consume the prediction and show the results. See the source code into frontend folder to predict-diseases application.


Top comments (0)