Reduce the time from research to production using various MLOps tools and techniques.
Refactoring a project developed in a notebook to a production-ready state can be challenging and consume our time and resources. At DagsHub, we’ve interviewed data science practitioners and ML engineers from hundreds of companies, trying to get to the bottom of their workflow problems and solutions. In this blog, I will construct our two years of research into a structured workflow that will help YOU reduce the iteration time between research and production using MLOps tools and techniques.
Considerations When Deploying a Model to Production
Implementing DagsHub’s product playbook, I’d like to start our journey at the end zone by thinking about our considerations when deploying a model to production.
When I think about production, the first thing that comes to mind is delivering a product to a different team — in or outside the company. Therefore, we’d like our results to be reliable, and in many cases, explainable. We’d also like to reduce the risk of bugs in production; therefore, our project should be testable, not only code but also model, data, pipeline, etc. When a bug arises in production, and it will, we need a way to reproduce the production’s state. This means not only having access to all different components but also the relevant versions. Last, we’d like the deploying process to be as automatic as possible, reducing the friction between the research team and the MLOps team.
Based on those considerations, let’s go back to the starting point. We’ll usually start a project by testing and prototyping our hypothesis, and our go-to tool will be Notebooks.
Project Jupyter
Born out of IPython in 2014, Project Jupyter has seen enthusiastic adoption among the data science community to become the de facto standard tool for prototyping. Its impact was officially acknowledged in 2017 when chosen to receive the ACM Software Systems Award, an honor shared with Unix, the Web, Java, and such.
However, if you explore some of the data science communities, you’d get the impression that notebooks are the root of all evil. I disagree. I think that Notebooks are awesome! They give us an interactive way to work with our code. Many companies, DagsHub included, choose the notebook interface to host their tutorials and interactive manuals. They have built-in visualizations, a benefit for communicating your work with technical colleagues or with not-so-technical stakeholders. All of this means they are great for quick prototyping.
The Downside of Using Notebooks in Production
However, when your project evolves and grows in complexity, not to mention moving to production, you most likely hit walls. Based on our research, we mapped five core difficulties data scientists face:
Reproducibility — With notebooks, we can run out-of-order code and edit the cells after they have already been run once. A huge disadvantage for reproducibility, which makes us put a lot of effort into tracking the kernel state when executing an experiment.
Version Control — Jupyter notebooks are basically large JSON files that can’t easily be diffed by Git, which makes it hard to review (not to mention merge) changes.
Debug — Notebooks have a debugger tool, but as the notebook gets longer, it becomes a real pain to use.
Testing & Reusing — The code hosted in notebook cells is not callable from external locations and can’t be tested easily.
CI/CD — Lacking proper CI/CD tools to automate the deployment process.
If you encountered other challenges while working with Notebooks — I’d love to hear about them!
Some even go as far as deploying the notebook to production, which in my opinion, is not recommended. It disregards the industry standards, workflows, and best practices. Also, we’re still missing proper CI/CD tools to support the deployment lifecycle, the notebook requires significant cleanup and packaging of libraries outside of the data scientist skillset, and it’s hard to run while using other tools in production.
“But hi, Netflix deploys their notebook to production”, an argument used a lot in the MLOps.community. Well, yes, some great companies use notebooks in production, but they invest a lot of resources to support such workflows, which in most cases you’ll not have or want to deflect for this task.
Components of a Solution
Having the above challenges and production considerations in mind, I gathered the best practices currently used in the industry into a structured workflow based on six components.
Convert to Scripts — convert the code into functions hosted in scripts.
Monorepo Strategy — Use the “monorepo for data science” structure to reduce complexity and scale our work.Version EVERYTHING — version all of the project components — code, data, model, etc.
Experiments Tracking — automate the logging process of experiments to aid our understanding of the project history and enable people to review the high-level results.
Export Logic — move the logic steps to external scripts to avoid using notebooks in production and maintain one codebase.
Unit Testing — Write tests for the project components, and invest more efforts in testing components that are low level and form the basis for other components.
Example scenario
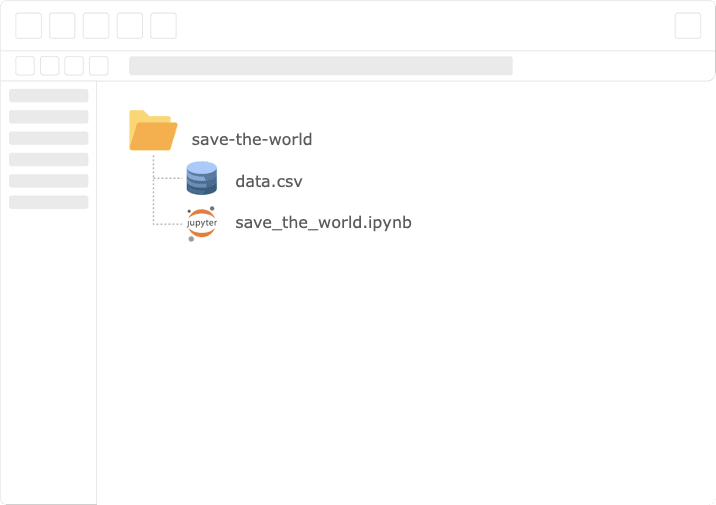
In our example scenario, the team lead just got off the phone with the board, and there is a new project in the funnel — “Save The World”. They just sent the dataset, and obviously — they want a model in production ASAP! So we open a new notebook and get our feet wet.
Convert to Scripts
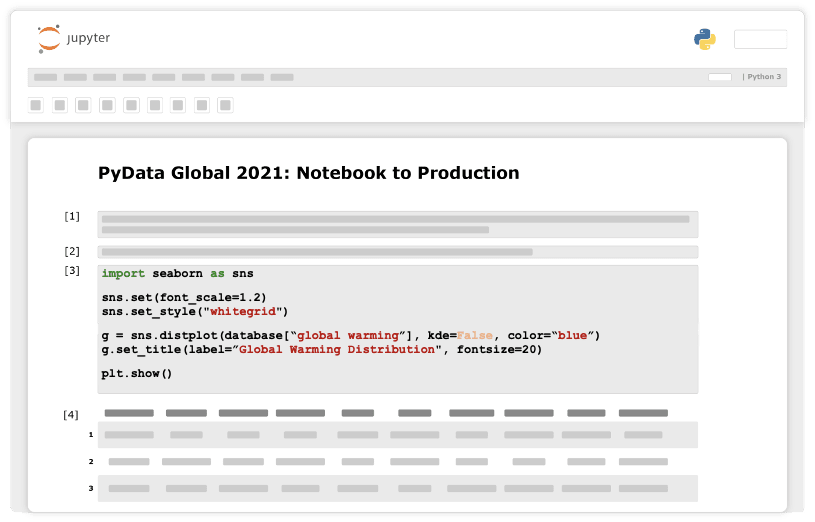
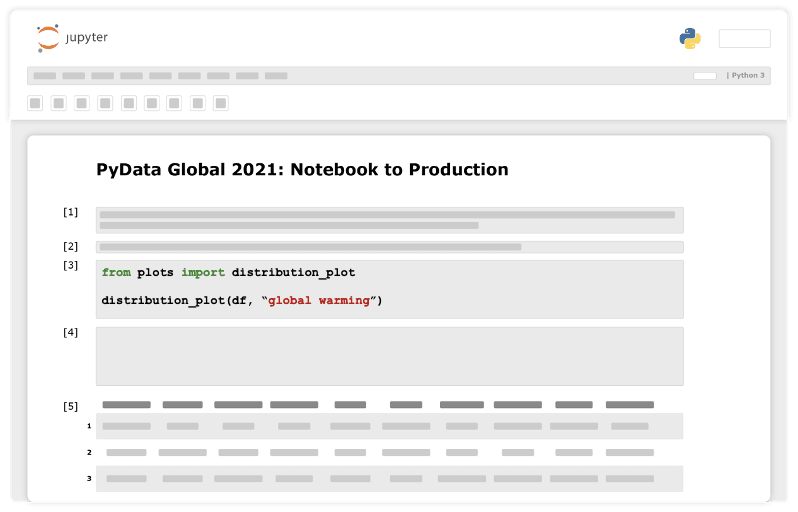
Our initial step will probably be exploring the data at hand. For example, check for feature distribution.
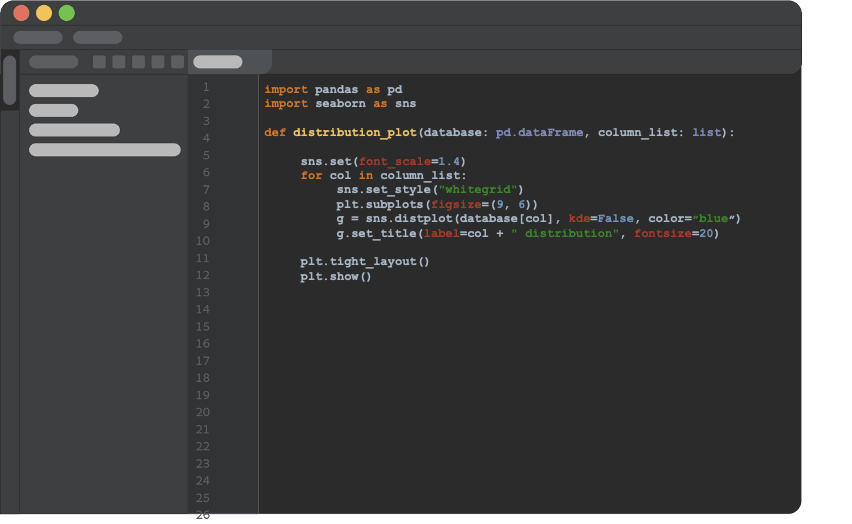
Now comes the most important conceptual change we need to make. After getting an initial piece of code to do what It was designed to do, we refactor it into a python function that doesn’t rely on any global variables and takes everything as arguments. We will store the function in python script, outside the notebook, which will make it callable from anywhere within the project.
Next, we will import the function to our notebook and use it.
Capabilities Unlocked
Code versioning — Version control our code using standard tools.
No hidden state — The function’s inputs and outputs are clearly defined with no side effects. The code is cleaner, testable, and repeatable.
Stay DRY (don’t repeat yourself) — Reuse the code in this project or anywhere else.
Lint — Use static code analysis tools, an industry-standard, to flag programming errors, bugs, stylistic errors, and suspicious constructs.
Monorepo strategy
Now that we made our project much more modular, we’d like to leverage it and become more scalable by implementing the good old monorepo strategy. Monorepo (“mono” meaning ‘single’ and “repo” is short for ‘repository’) is a trunk-based development implementation where code for many projects is stored in the same repository. Well-known companies use this strategy, such as Google, Facebook, Microsoft, and more.
What has monorepo got to do with Data Science?
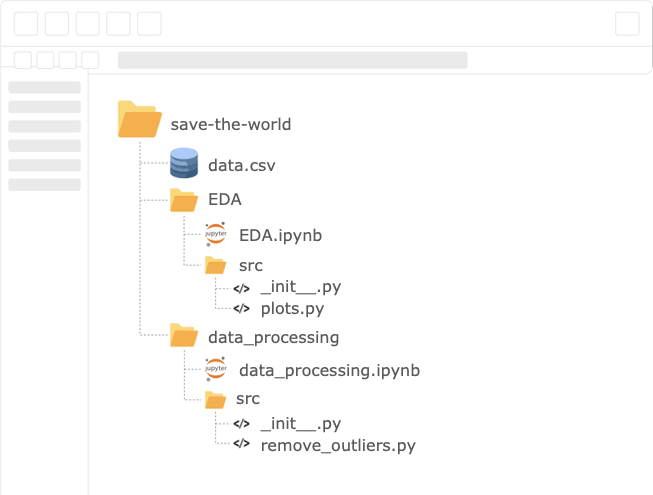
Data science projects can be divided into stages that are completely independent of the other, e.g., EDA, data processing, modeling, etc. If we address each step as a separate component, we will be able to divide the project into sub-tasks that are actually sub-projects. Using this mindset, we’ll be able to scale our work by dividing the tasks between different team members that will be independent of one another and conquer the project simultaneously.
We will notice that each stage depends only on the output of the previous one. For example, the modeling step only needs the output of the processing stage. This is why we’d like to open the bottleneck by producing a minimum valuable pipeline, MVP, and have each sub-team start working on its task. Once a team reaches a useful product, they release an update of their output.
Now, we’ll have a python module for every sub-task that stands on its own and as an integral component in the project’s pipeline. Moreover, Instead of having one long notebook for the entire project, we’ll have a notebook per task.
Capabilities Unlocked
Scale — We can scale the number of collaborators on the project, increase our [product/time], and move X times faster to production.
Maintain, review & debug — Having shorter notebooks, that are task-oriented, will make it easier to maintain, review & debug them.
Onboarding — The clear structure of the project makes the onboarding process easier.
Version Control
Once reaching a point where we’re collaborating with other people, it rings only one bell — Version Control. For this task we’ll use Git, an open-source tool widely adopted in the industry, that is great for modeling parallel work and versioning code files. Git enables us to use standard CI/CD tools, recover previous work with a click of a button, insulate the working environment, and more. If you want to dive into Git for Data Science, Martin explains it perfectly in his post.
But what about the non-code artifacts?
Let’s say that the data processing team created a new data set; should they just overwrite the old version? Or maybe stone-age-version it by creating a new folder and giving it a meaningful name (e.g., new_data_v3_fixed_(1)_best.csv)? Well, both options are inadequate. This is why we’d want to also version our large files.
Git was designed for managing software development projects and for versioning text/code files. Therefore, Git doesn’t handle large files. Git released Git LFS (Large File System) to overcome large file versioning, which is better than Git, but fails when scaling. Also, both Git and Git LFS are not optimized for data science workflow. To overcome this challenge, many powerful tools emerged in recent years, such as DVC, Delta Lake, LakeFS, and more.
At DagsHub, we’re integrated with DVC, which I love using. First and foremost, it’s open-source. It provides pipeline capabilities and supports many cloud providers for remote storage. Also, DVC acts as an extension to Git, which allows you to keep using the standard Git flow in your work. If you don’t want to use both tools, I recommend using FDS, an open-source tool that makes version control for machine learning fast & easy. It combines Git and DVC under one roof and takes care of code, data, and model versioning. (Bias alert: DagsHub developed FDS)
Capabilities Unlocked
Reproducibility — Having the state of all the project components under one version enables us to reproduce results with a click of a button.
Avoid ad-hoc versioning done by filenames. The code outputs to the same file and versions it.
Avoid overwriting work because of small mistakes and have the ability to recover files easily.
Sharing & synchronizing versions make collaborating with others easier.
Experiment Tracking
Moving our code to functions makes it easier to control the input they get. This helps us manage the configuration of every run and track the hyperparameters of each experiment easily. We’ll move the configurations to an external file, and each module will import its relevant variables.
But as you can imagine, tracking each experiment with Git can become a hassle. We’d like to automate the logging process of each run. The same as for large file versioning, many tools emerged in recent years for experiment logging, such as W&B, MLflow, TensorBoard, and the list goes on. In this case, I believe that it doesn’t matter with which hammer you choose to hit the nail, as long as you punch it through.
Note that when using external tools, you only log the experiment information without the ability to reproduce it. Therefore, I recommend Mix n’ Match between the tool and the use case. Prototyping a new hypothesis will probably not provide meaningful results; therefore, we’ll use the external tool to log the experiment. However, after reaching a meaningful result, we’ll version all of the project components and encapsulate them under a Git commit, making it reproducible with a click of a button.
Capabilities Unlocked
Research history — Automating the experiment tracking process will assure logging of all the research history, enable us to understand the high-level results, and avoid repeating our work twice.
Sharing — Share the experiment results with the team while having the ability to compare apples to apples.
Reproducibility — reproduce the experiments (to some extent.)
Avoid ad-hoc recording of experiments, which will vary between the team members.
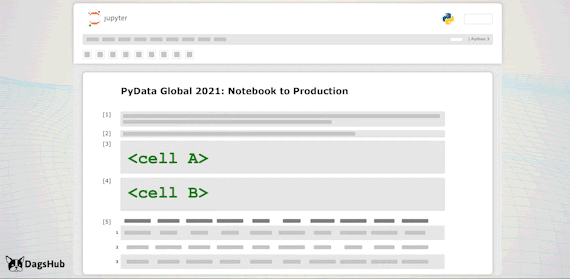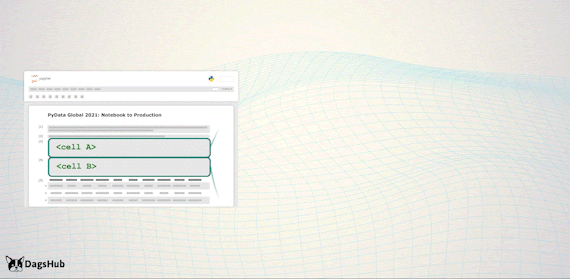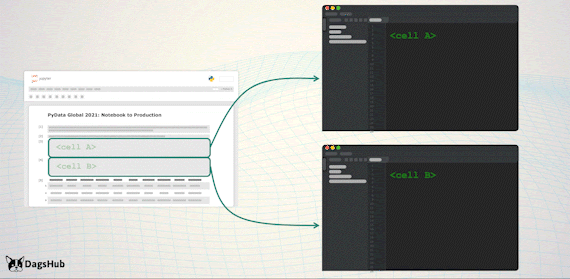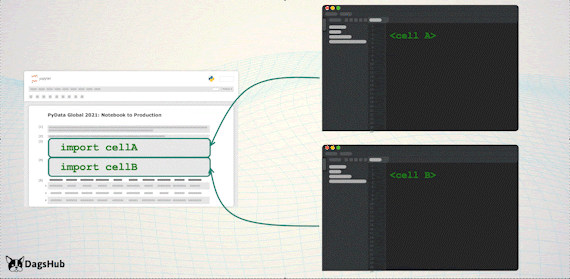
Export Logic to Scripts
Just before we deploy to production, we’d like to move all of the logic steps stored in the notebook, to python scripts. However, we wouldn’t want to lose the notebook capabilities; therefore, we’ll import them back to the notebook.
Capabilities Unlocked
Use only scripts in production, which is the industry standard, and much easier to handle.
Maintain one code base and not in both notebook and scripts.
Keep using notebooks and utilizing their advantages.
Unit Tests
The same as in software development, we’d want to make sure that our code/model does whatever it was designed to do. It goes with data processing, modeling, and all pipeline stages. Writing tests during coding actually makes you work faster overall. Since you get a tight fast feedback loop that catches bugs early, it’s much easier to debug problems since you can debug each unit in isolation instead of debugging a whole running system.
Capabilities Unlocked
- Fewer bugs in production = More sleeping hours.
Summary
Jupyter notebook is an excellent tool for prototyping and exploratory analysis, that after all, gives us superhuman abilities. However, when working in a production-oriented environment, its limitations are revealed. When a new project comes in the funnel, I highly recommend thinking about the production considerations, what tools should be used in the project, and how you can make your work more efficient. You might measure your work by the model’s performance, but you are measured by the iteration time to, and quality in, production.
If you’re facing challenges that I didn’t mention in this post or thinking of better ways to overcome them — I’d love to hear about it! Feel free to reach out on LinkedIn and Twitter.




Top comments (2)
Thanks for writing this. Love python, Love Jupyter, needed a point of reference on a few things, this post covered all the bits I was unsure of. Well written!
Thanks for this amazing article.